Prost!
Sequencing Analysis Known miRNA Identification IsomiRs Identification
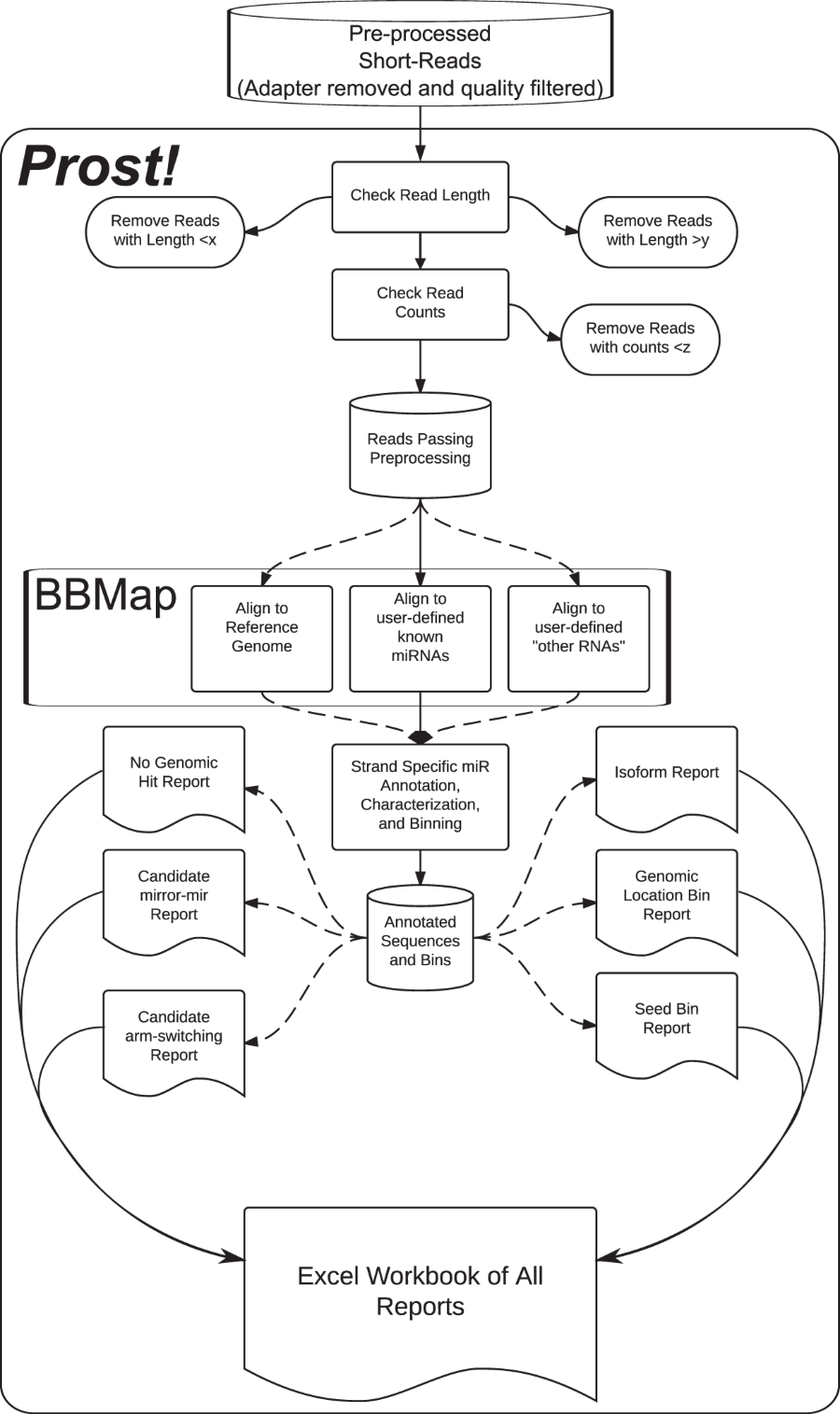
Prost! (PRocessing Of Short Transcripts) analyzes any source of smallRNA sequencing data. Prost! does not rely on existing annotation to filter sequencing reads but instead starts by aligning all the reads on a user-provided genomic reference, allowing the study of miRNAs in any species. Additionally, any number of samples can be studied together in a single Prost! run, allowing a more accurate analysis of an entire dataset. After grouping the processed reads by genomic location, Prost! then annotates the reads using a user-defined annotation database (public or personal annotation database). Genomic alignment, grouping, and then annotation enable the study of potentially novel miRNAs, as well as permitting the retention of all the isomiRs that a miRNA may display. Finally Prost! contains additional features such as grouping by seed sequence for a more functional approach of the dataset, provides automatic discovery of potential mirror-miRNAs, and analyzes the frequency of various types of post-transcriptional modifications at each genomic location. Each step of the Prost! analysis are provided into the excel file output for the user to have all information in hands for deeper analysis of specific cases.
Homepage: Link
Last Software Update: March 11, 2019
Publication Date: March 8, 2019
Citations: 23 [via Pubmed]
Organism Specific:
Reference Genome Needed:
Online/Local:
/
Installation/User Level: intermediate
User Adjustability:
User support:
Precomputed Target Results Available For Download:
License: FLOSS
Input Data Required: