SomamiR DB 2.0
miRNA Databases
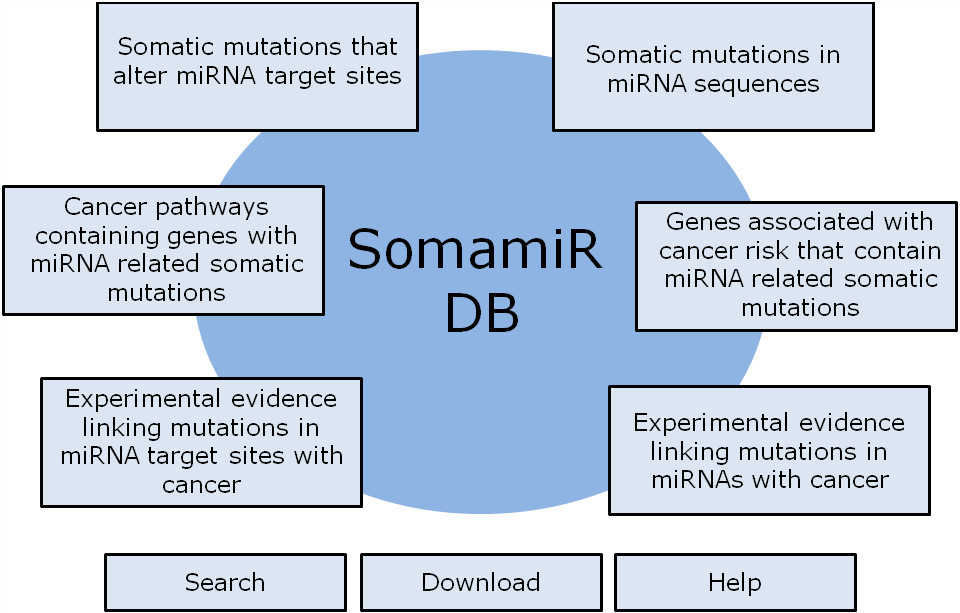
SomamiR is a database of cancer somatic mutations in microRNAs (miRNA) and their target sites that potentially alter the interactions between miRNAs and competing endogenous RNAs (ceRNA) including mRNAs, circular RNAs (circRNA) and long noncoding RNAs (lncRNA). It also provides an integrated platform for the functional analysis of these somatic mutations. Additionally, the SomamiR DB contains a collection of somatic and germline mutations in miRNAs or their target sites that have been strongly linked with cancer. Specifically, the majority of these mutations met the following criteria (a) they have been associated with cancer risk in GWAS or CGAS and (b) there is experimental evidence indicating that the mutation alters miRNA function
Homepage: Link
Last Software Update: Nov. 27, 2015
Publication Date: Nov. 17, 2015
Citations: 71 [via Pubmed]
Organism Specific:
Reference Genome Needed:
Online/Local:
/
Installation/User Level: easy
User Adjustability:
User support:
Precomputed Target Results Available For Download:
Cross-Refferences: